Changing Paradigm
Conventional biochemistry mentality is based on specific structures and specific interactions. This way of thinking has numerous advantages because using it allows one to crystalize and investigate proteins, to see molecules moving together (electrophoresis), to crosslink interacting partners etc. and thus deduct molecular mechanisms. However, the paradigm starts to shift. A large fraction of proteins and protein domains appear to be intrinsically disordered (by different estimates 30-50% of the proteome), proteins are engaged in multiple and fuzzy interactions, and the real crowded and messy environment of the cell appears to be far different than the pure lonely space of the in vitro world.
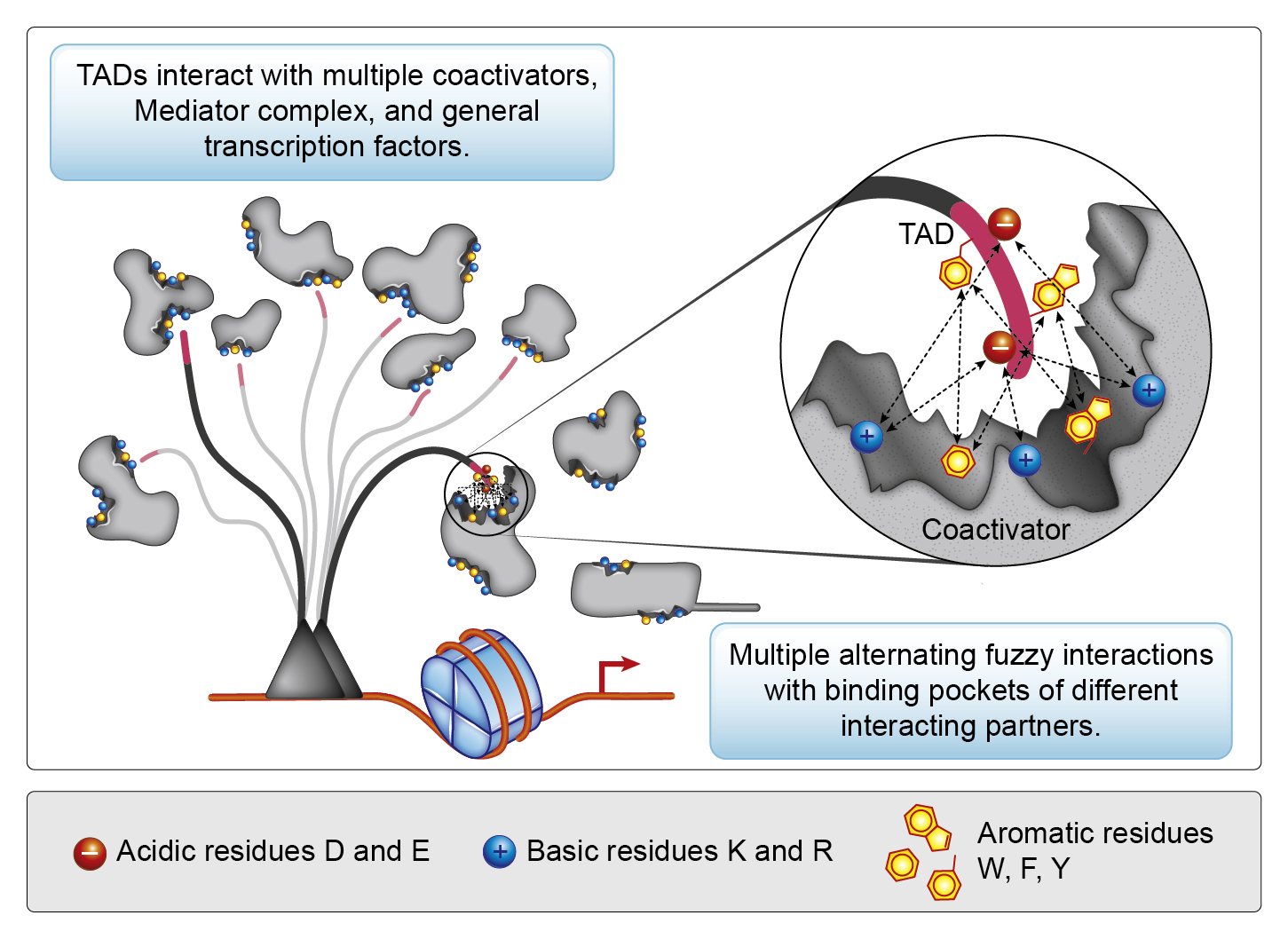
The transcriptional activation domains (TADs) fully represent the new paradigm. For several decades we thought of TADs as peptides with a specific or semi specific (induced fit) structure, specific or semi specific consensus sequence, interacting with a specific target or a set of transcription related targets. Using this logic we struggled for decades to understand the mechanism of TAD function and failed. The molecular biology textbooks talk little about TADs and the brief paragraphs usually mention a model based on conventional biochemistry of (semi)specificity, which starts to crumble if applied to the real near-stochastic world of TADs.
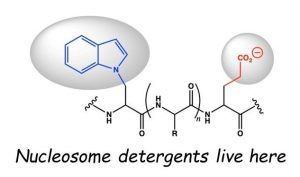
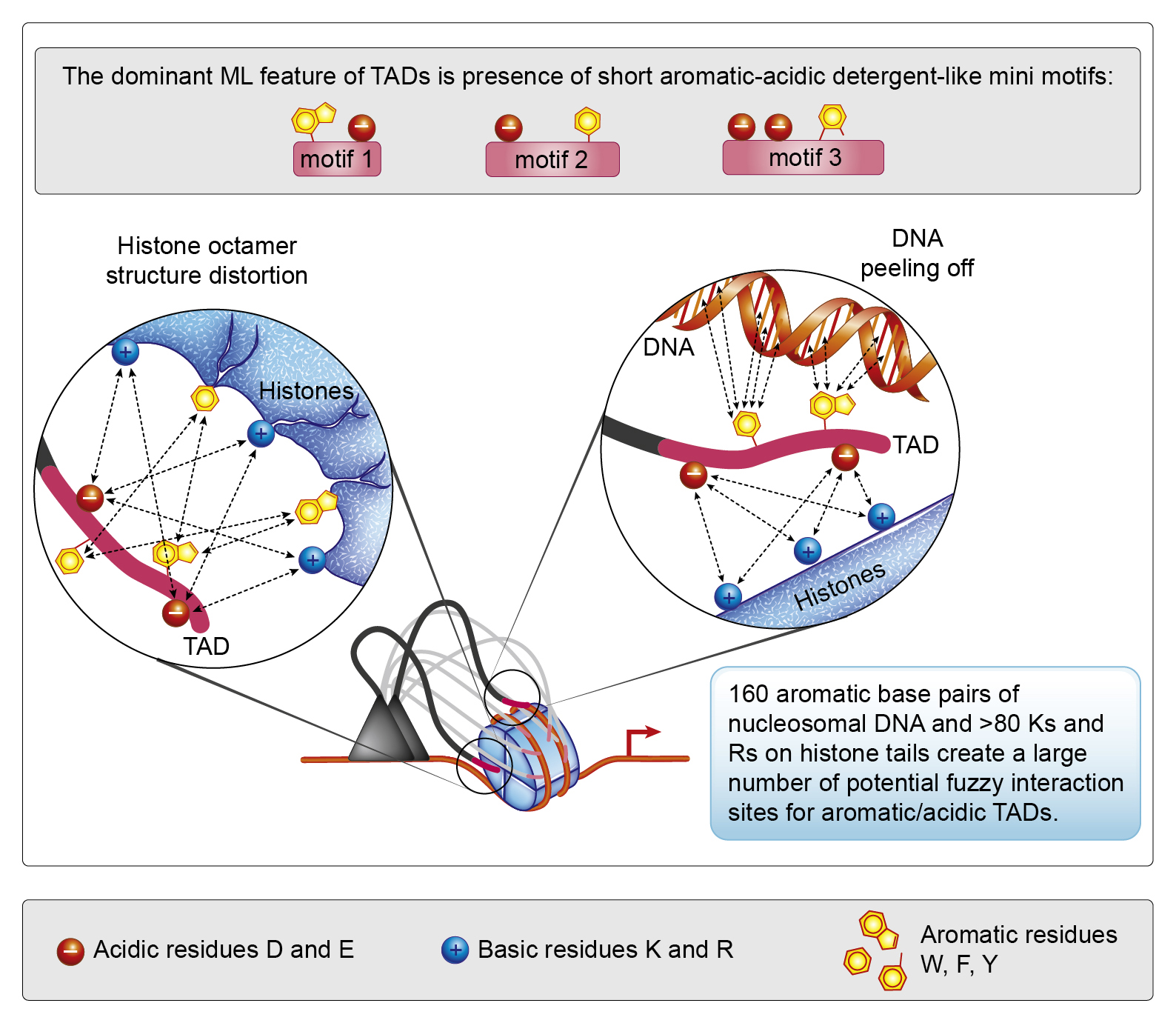
We propose a different model for TAD function based on the nucleosome detergents concept. It explains multiple contradictions and uncertainties perfectly, however the biggest challenge is to convert the mentality of specificity into the mentality of multiplicity in the crowded near-stochastic world of the cell. This is akin to switching from thinking about falling objects as metal balls to thinking about them as falling autumn tree leaves. Analogous to nonlinear physics, we call this new thinking framework – nonlinear biochemistry.
BTW, adopting nonlinear biochemistry means also adopting new tools such as machine learning, statistics of large sequence pools, analysis of large cellular communities, and high throughput experimentation.